Reference Browsers
QiTissue provides an easy-to-access reference hub for information on biomarkers, cell or image features, and an additional browser for access to common databases. They can be displayed in tandem with your workflow, and in certain instances edited to accommodate visual or analytical preferences.
Web Reference Browser
The Web Reference Browser allows you to surf the web when working within QiTissue, with quick access to links for application documentation and tutorials, common databases for gene, cell, and protein information, and more.

Feature Reference
The Feature Reference window allows you to search any cell or image-based analytical feature. Just scroll through the lists or type in the search filter.
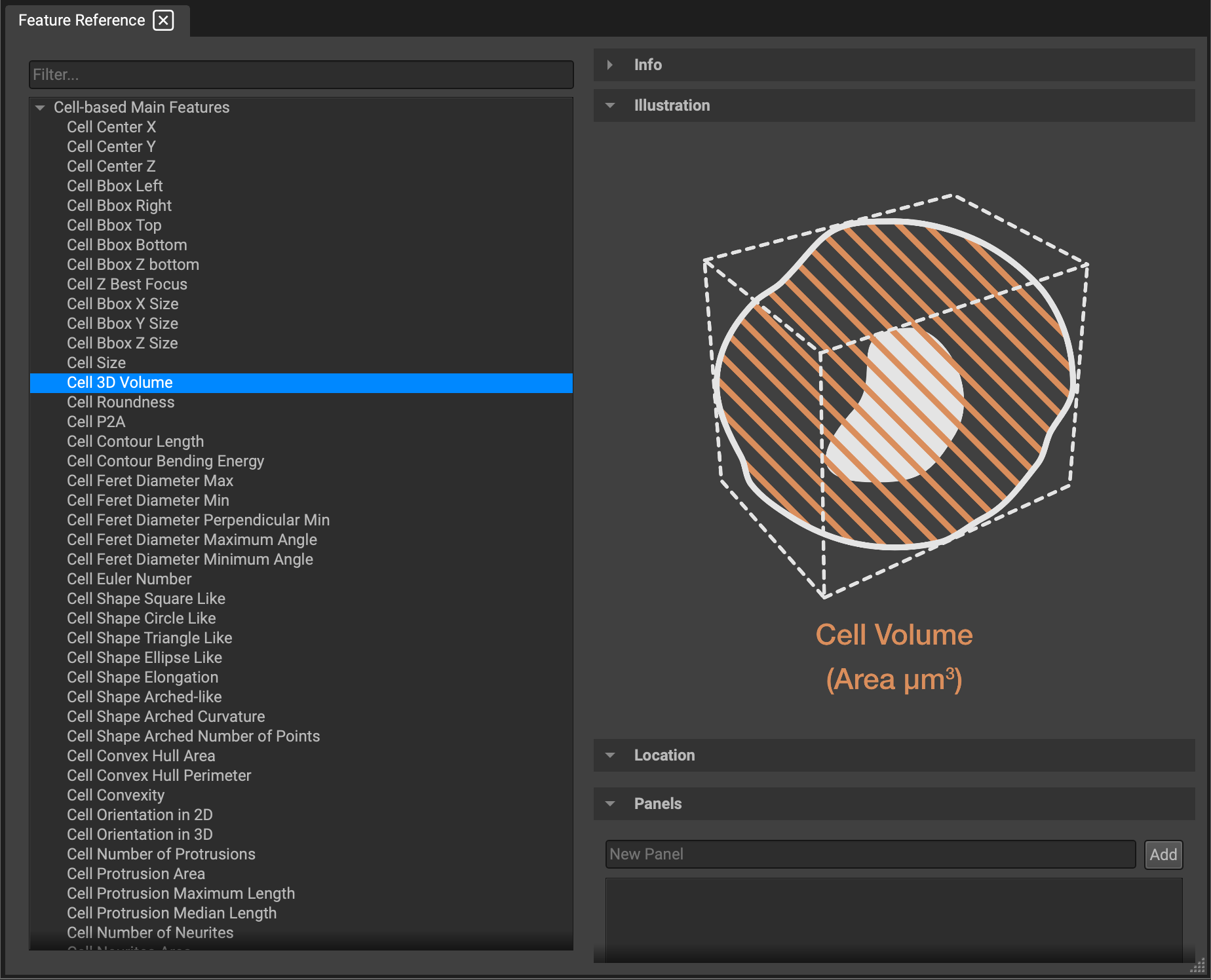
Currently, QiTissue computes feature extraction for roughly 70 image-based features and 20 cell-based features. However, the reference library contains over 800 features in total, including additional features users would like to import for analysis. Though these are not yet readily available to compute and use within QiTissue, they will be in the near future. (Note: this widget is still under construction).
Biomarker Reference
The Biomarker Reference window provides access to a wealth of knowledge on biomarkers within our library. You can retrieve, as well as locally edit, a lot of information; defining characteristics, staining patterns, relevant cancer hallmarks, cell type identifiers, segmentation and analysis presets, panels, and more.
The information is organized in the form of tabs along the right side of the window. You can filter through categories of biomarker (Architecture, Cell State, Immune, Cluster of Differentiation, and Immune Checkpoints) along the left side of the window. The search bar also provides a quick way to look up a particular biomarker.
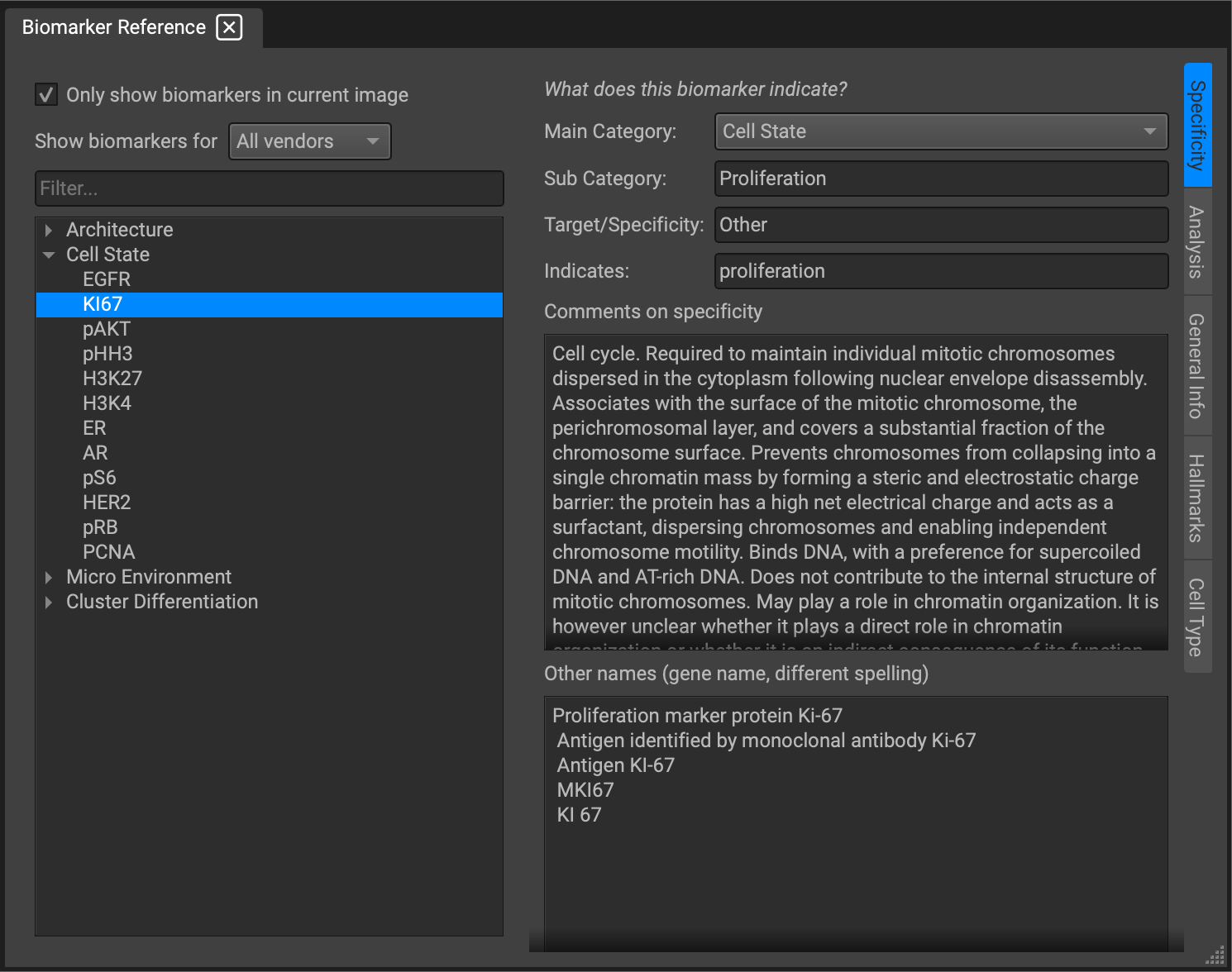
Information programmed into this space is helpful for a variety of reasons. For example, it can become time intensive to finetune the display settings for each of your biomarkers on an image. With this information stored in the analysis section of the widget for 1000+ biomarkers built into the program (including any additional markers you may like to add), QiTissue will load these in exactly as specified. This makes the workflow experience much more user friendly. Is the biomarker in the nucleus, cytoplasm, or cell surface? What value does it bring to the analytics? Is it just measured for expression, or is it an RNA marker for counting spots? Is texture important? Or intensity? All of this can be specified in this knowledgebase.
The general info tab also contains shortcut buttons to websites for vendor catalogs or UniProt protein information, which will immediately pull up the web reference browser.
Also known are specificities for cancer hallmarks cell types, and their +/- relationships to these cell types. Built-in heuristics automatically predict which biomarkers you will want to visualize, such as cell death or proliferation markers, and can suggest certain cell type markers to display side by side. Eventually, QiTissue will actively suggest color combinations and sequencer movies that can save you a lot of time, given how many permutations of colors are possible when exploring.

